Instructions for Installing and Executing the Triaxus R script on a windows machine
Steps
Install the latest version of R from http://cran.r-project.org/bin/windows/base/ with default settings. Under the default settings, R should be installed in the "C:\Program Files\R" directory.
Run R in interactive mode by executing R.exe as an Administrator. It would be available at "C:\Program Files\R\R-3.1.1\bin\R.exe" and install the various necessary packages by entering the following commands in the command prompt:
install.packages("ggplot2",dependencies=T)install.packages("gstat",dependencies=T)
install.packages("moments",dependencies=T)
install.packages("fields",dependencies=T)
install.packages("GA",dependencies=T)
install.packages("spdep",dependencies=T)
install.packages("rgeos",dependencies=T)
install.packages("fields",dependencies=T)
install.packages("gWidgets",dependencies=T)
install.packages("gridExtra",dependencies=T)
install.packages("dismo",dependencies=T)
- Download the code from https://uofi.box.com/s/p970k1fz83gz5cyn3n3h Extract the zipped folder and you should be able to see the contents as shown below.
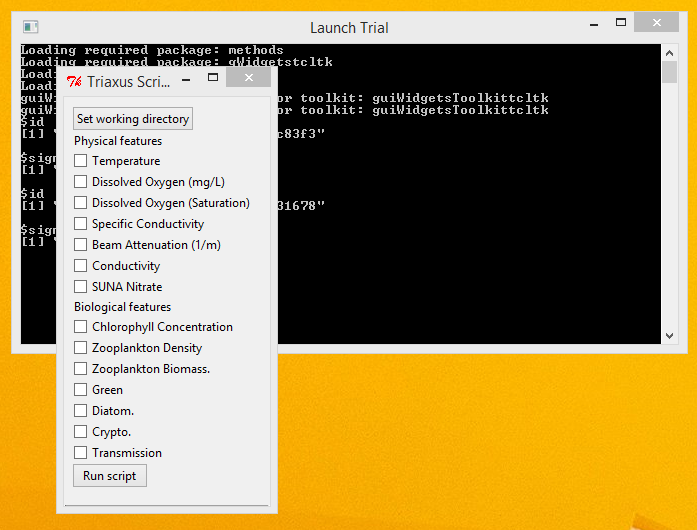
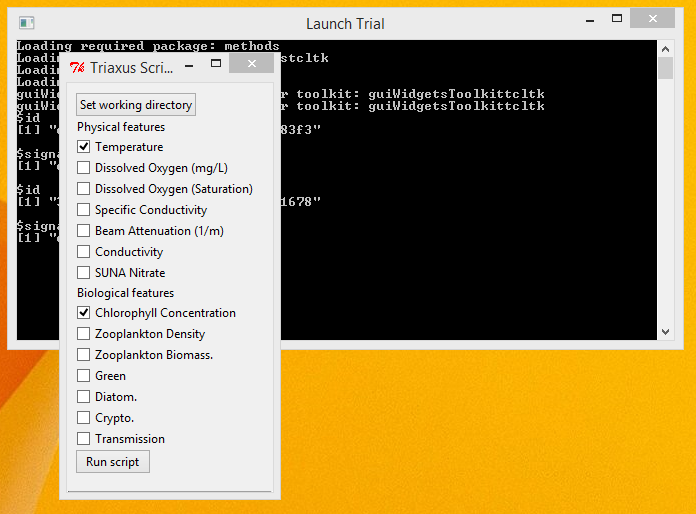
Execution - Execute the application by double clicking the 'Launch Trial' shortcut. You will see an interface as shown below.
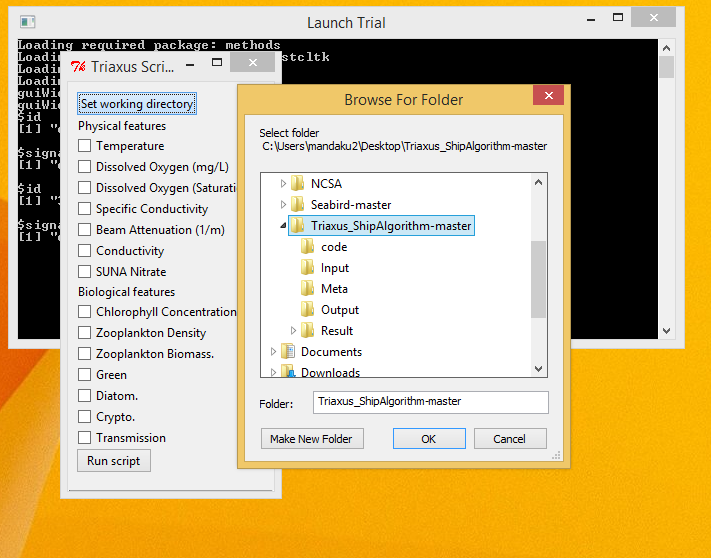
- Select the main directory for the application where all the files exist as the working directory, by clicking on the Set working directory button.
- Copy all the data files to the 'Input' folder of the Triaxus script directory.
- Select the various Physical and/or Biological features that you want the script to consider, by checking the appropriate boxes adjacent to each of the features.
- Click the 'Run script' button to start the script execution.
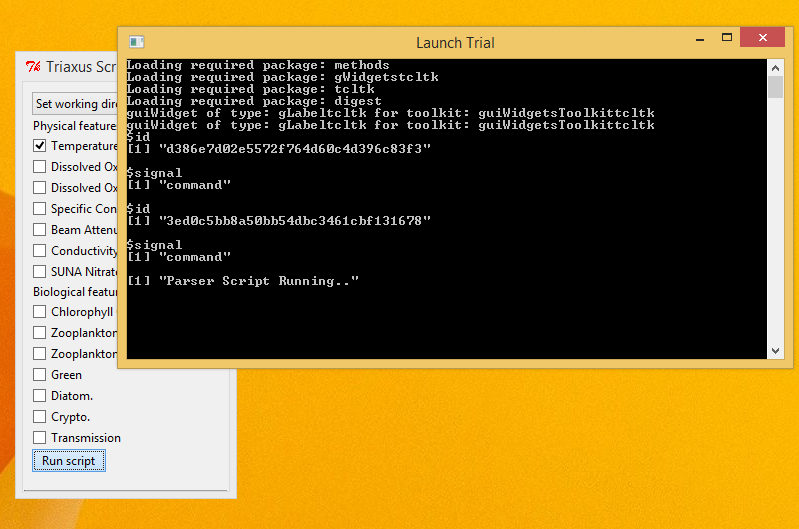
- The Console window would give real time status updates on the script's execution.
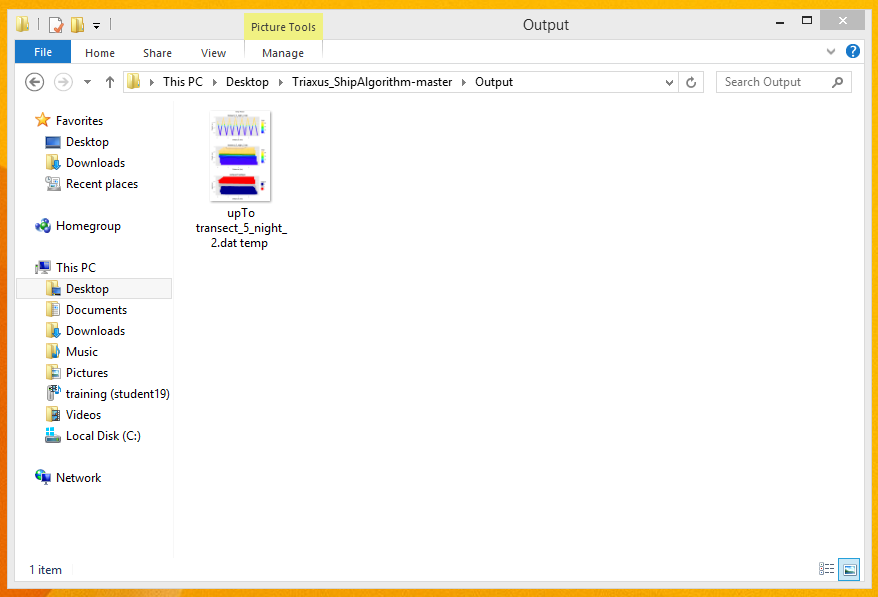
- Once the script finishes execution, the output files should have been generated inside the 'Output' folder in the Triaxus directory.
- Sample Outputs generated from the Triaxus Script for the Temperature and SUNA Nitrate features Triaxus Script Output : Feature = Temperature
Instructions for running the Seabird script on a windows machine
Steps
- Install Python version 2.7 from https://www.python.org/download/releases/2.7.7/
Install the scikit-learn machine learning library available as "scikit‑learn‑0.15.1.win32‑py2.7.exe" at
http://www.lfd.uci.edu/~gohlke/pythonlibs/#scikit-learnInstalling the Scipy stack for python 2.7 available as "Scipy-stack-14.5.30.win32-py2.7.exe" at
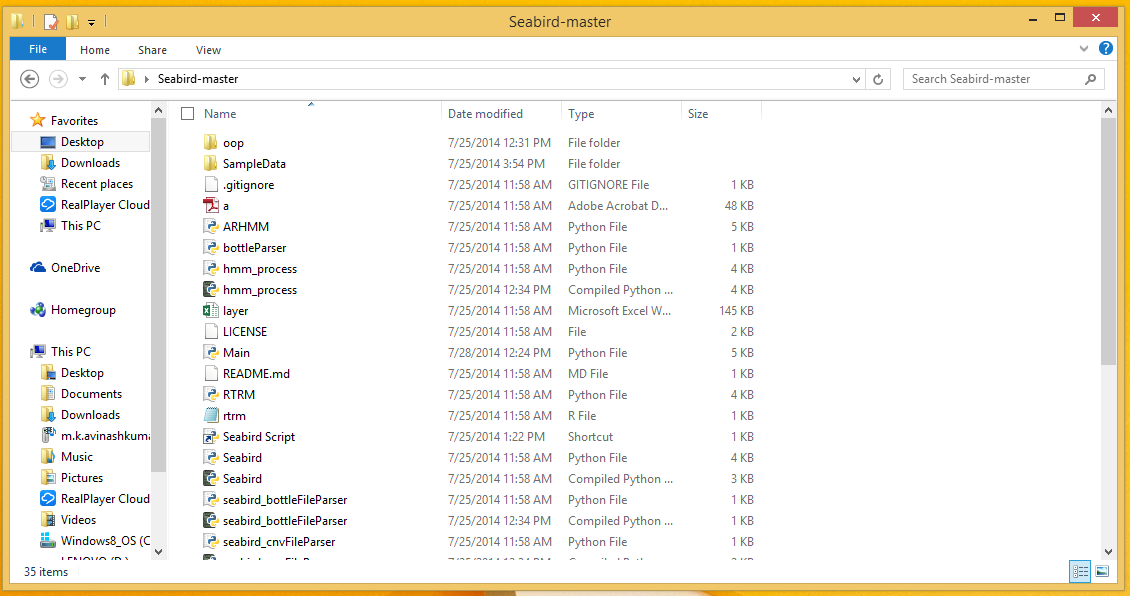
http://www.lfd.uci.edu/~gohlke/pythonlibs/#scipy-stack- Download the code from https://uofi.box.com/s/elmzg038rmyv16v8yht5 Extract the zipped folder and you should be able to see the contents as shown below.
- Execute the application by double clicking on the 'Seabird Script' shortcut.
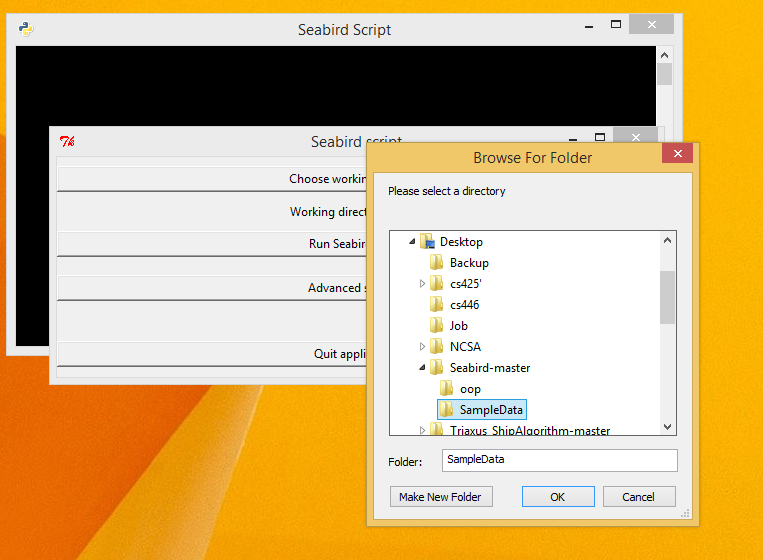
- Choose the directory where all the input data files as the working directory by clicking on the 'Choose working directory' button.
- If you wish to change some advanced settings for running the script, click on the 'Advanced Settings' button.
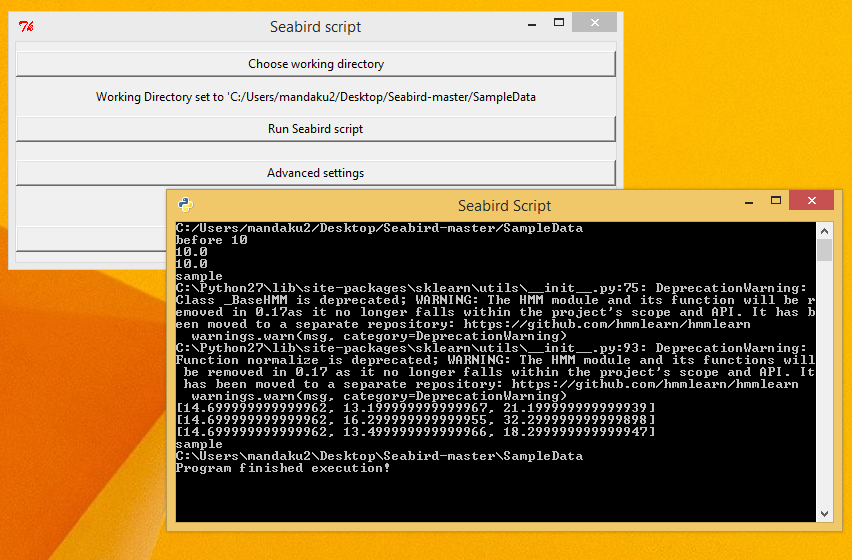
Click on the 'Save Settings' button to save the changes and get back to the main page. - Choose the 'Run Seabird Script' button to execute the script with the settings previously chosen.
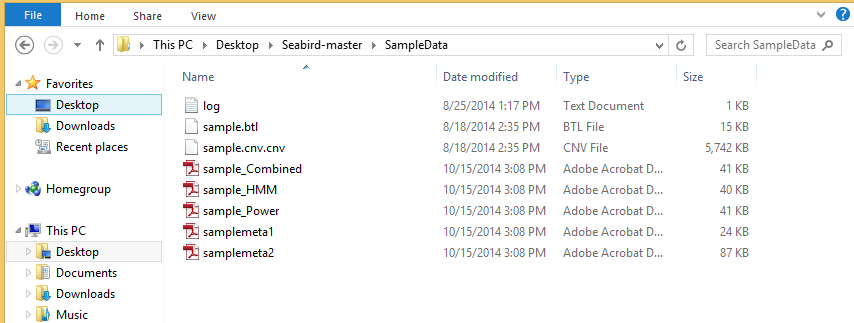
You should be able to see updates on the command line window as shown. - The output would have been generated in the 'SampleData' folder inside the Seabird script directory.
- Sample outputs generated from the Seabird Script HMM & Power Spectra Model Combined Output
Instructions for running the Triaxus R script on a linux machine
Steps
- Install R
- sudo apt-get install r-base
- sudo apt-get install libgeos++-dev
- Go to the R execution environment by entering sudo R
- Install various prerequisite packages through the following commands
install.packages("ggplot2")
install.packages("gstat")
install.packages("moments")
install.packages("fields")
install.packages("GA")
install.packages("spdep")
- Create a Folder called Result inside the applications directory and a sub folder called Variogram inside the newly created Result folder. These folders would be used by the script to store some meta data.
- Run the main entry R script for the application, by entering source("Main.R")
- The final output graph should be available as a pop up on the terminal.
Instructions for running the Triaxus R script on a linux machine
Download the seabird code from https://github.com/stormxuwz/Seabird
Steps
- Install the latest version of Python
- Install Python Package Manager
- sudo apt-get install python-pip
- Install all the required packages in the SciPy library stack
- sudo apt-get install python-numpy python-scipy python-matplotlib ipython ipython-notebook python-pandas python-sympy python-nose
Install the scikit learn library for python
sudo pip install -U scikit-learn
- The main entry program for this application is the Seabird.py python script
- In the script Seabird.py in lines 89,101 specify the directory under which the Seabird input data files are present inside the os.chdir command. In line 102, specify the name of the .btl file which contains the seabird bottle data in the variable bottle_file.
- Save and run the script.
- python Seabird.py
- The output files would have been generated inside the directory which you previously specified in step 6.