http://sc16.supercomputing.org/ - Wednesday, Nov. 16th, 3:00pm MST
Goals
- NDS Share(d) datasets: present online the below datasets so that users can find and obtain them. Highlight DOI's of both paper and dataset.
- Provide analysis tools along with each dataset (ideally deployable data-side to avoid having to copy the data).
Presentation Logistics
- Booth demos: NCSA, SDSC, ...
- https://docs.google.com/spreadsheets/d/1WXtu0wpU5bemFKEIfbYRmkasw_w5Po4epiy4tiLjkJc/edit#gid=0
- Wednesday, Nov. 16th, 3:00pm MST
- Draft list of specific individuals to invite to booths
- Create a flyer (address how we approached the problem? discuss tech options)
- SCinet late breaking news talk
Technology
- Globus Publish (https://github.com/globus/globus-publish-dspace)
- Modified DSpace with Globus Authentication, Groups, and Transfer
- Example instance: Materials Data Facility
- Requires a Globus endpoint on each resource (not available for Swift stores, e.g. SDSC Cloud)
- Can skin a collection (currently doesn't support skinning instance)
- Analysis currently not supported
- yt Hub
- Built on Girder with additional plugins to support Jupyter, ownCloud, ...
- Example skinned instance: Galaxy Cluster Merger Catalog
- Would need allocation on BlueWaters to run tools there. May need to move Moesta dataset elsewhere...
- Resolution Service
- Given DOI → get URL to data, get loction, get machine, get local path on machine
- Given notebook, location, path → run notebook on data at resource
- Allows independence from repo technologies
- Allow repos to provide location information as metatdata, if not available attempt to resolve (e.g. from URL, index)
- Repos that don't have local computation options would need to move data
- Only requirement from repos is that data can be accessed via a URL
- Identify at least one notebook for demonstration
- Build as service with python library interface that can be shown in Jupyter
- Create an alternative bookmarklet client that can be shown on any of repo
- click on link get list of resources to run a selected notebook on
- Discussed as a need within TERRA effort
- Leverage work with Girder in Whole Tale:
Datasets
1. MHD Turbulence in Core-Collapse Supernovae
Authors: Philipp Moesta (pmoesta@berkeley.edu), Christian Ott (cott@tapir.caltech.edu)
Paper Citation: Mösta, P., Ott, C. D., Radice, D., Roberts, L. F., Schnetter, E., & Haas, R. (2015). A large-scale dynamo and magnetoturbulence in rapidly rotating core-collapse supernovae. Nature, 528(7582), 376–379. http://dx.doi.org/10.1038/nature15755
Paper URL: http://www.nature.com/nature/journal/v528/n7582/full/nature15755.html
Paper DOI: dx.doi.org/10.1038/nature15755
Data Citation: ??
Data URL: https://go-bluewaters.ncsa.illinois.edu/globus-app/transfer?origin_id=8fc2bb2a-9712-11e5-9991-22000b96db58&origin_path=%2F
Data DOI: http://dx.doi.org/doi:10.21970/N9RP4P
Data Location: Blue Waters (/projects/sciteam/jr6/share/)
Size: 205 TB
Code & Tools: Einstein Toolkit, see this page for list of available vis tools for this format
Jupyter Notebook: ??
The dataset is a series of snapshots in time from 4 ultra-high resolution 3D magnetohydrodynamic simulations of rapidly rotating stellar core-collapse. The 3D domain for all simulations is in quadrant symmetry with dimensions 0 < x,y < 66.5km, -66.5km < z < 66.5km. It covers the newly born neutron star and it's shear layer with a uniform resolution. The simulations were performed at 4 different resolutions [500m,200m,100m,50m]. There are a total of 350 snapshots over the simulated time of 10ms with 10 variables capturing the state of the magnetofluid. For the highest resolution simulation, a single 3D output variable for a single time is ~26GB in size. The entire dataset is ~205TB in size. The highest resolution simulation used 60 million CPU hours on BlueWaters. The dataset may be used to analyze the turbulent state of the fluid and perform analysis going beyond the published results in Nature doi:10.1038/nature15755.
2. Probing the Ultraviolet Luminosity Function of the Earliest Galaxies with the Renaissance Simulations
Authors: Brian O'Shea (oshea@msu.edu), John Wise, Hao Xu, Michael Norman
Paper Citation: Norman, B. W. O. and J. H. W. and H. X. and M. L. (2015). Probing the Ultraviolet Luminosity Function of the Earliest Galaxies with the Renaissance Simulations. The Astrophysical Journal Letters, 807(1), L12.
Paper URL: http://iopscience.iop.org/article/10.1088/2041-8205/807/1/L12/meta;jsessionid=40CF566DDA56AD74A99FE108F573F445.c1.iopscience.cld.iop.org
Paper DOI: dx.doi.org/10.1088/2041-8205/807/1/L12
Data URL:
Data Citation: ??
Data DOI: ??
Size: 89 TB
Code & Tools: Enzo
Jupyter Notebook: http://yt-project.org/docs/dev/cookbook/cosmological_analysis.html
In this paper, we present the first results from the Renaissance Simulations, a suite of extremely high-resolution and physics-rich AMR calculations of high-redshift galaxy formation performed on the Blue Waters supercomputer. These simulations contain hundreds of well-resolved galaxies at z ~ 25–8, and make several novel, testable predictions. Most critically, we show that the ultraviolet luminosity function of our simulated galaxies is consistent with observations of high-z galaxy populations at the bright end of the luminosity function (M1600 ⩽ -17), but at lower luminosities is essentially flat rather than rising steeply, as has been inferred by Schechter function fits to high-z observations, and has a clearly defined lower limit in UV luminosity. This behavior of the luminosity function is due to two factors: (i) the strong dependence of the star formation rate (SFR) on halo virial mass in our simulated galaxy population, with lower-mass halos having systematically lower SFRs and thus lower UV luminosities; and (ii) the fact that halos with virial masses below ~2 x 10^8 M do not universally contain stars, with the fraction of halos containing stars dropping to zero at ~7 x 10^6 M . Finally, we show that the brightest of our simulated galaxies may be visible to current and future ultra-deep space-based surveys, particularly if lensed regions are chosen for observation.
3. Dark Sky Simulation
Authors: Michael Warren, Alexandar Friedland, Daniel Holz, Samuel Skillman, Paul Sutter, Matthew Turk (mjturk@illinois.edu), Risa Wechsler
Paper Citation: Warren, M. S., Friedland, A., Holz, D. E., Skillman, S. W., Sutter, P. M., Turk, M. J., & Wechsler, R. H. (2014). Dark Sky Simulations Collaboration. Zenodo. https://doi.org/10.5281/zenodo.10777
Paper URL: https://zenodo.org/record/10777#.V_VvKtwcK1M, https://arxiv.org/abs/1407.2600
Paper DOI: http://dx.doi.org/10.5281/zenodo.10777
Data Citation: Warren, M. S., Friedland, A., Holz, D. E., Skillman, S. W., Sutter, P. M., Turk, M. J., & Wechsler, R. H. (2014). Dark Sky Simulations Collaboration. Zenodo. https://doi.org/10.5281/zenodo.10777
Data URL:
Data DOI: https://doi.org/10.5281/zenodo.10777 (Although this is classified as a report in Zenodo, the authors intended this to be the DOI for the dataset)
Size: 31 TB
Code & Tools: https://bitbucket.org/darkskysims/darksky_tour/
Jupyter Notebook: https://girder.hub.yt/#user/570bd8fc2f2b14000176822c/folder/5820b9c09ea95c00014c71a1
The cosmological N-body simulation designed to provide a quantitative and accessible model of the evolution of the large-scale Universe.
4. ...
Design Notes
Planning discussion 1 (NDSC6)
Photo of whiteboard from NDSC6
- On the left is the repository landing page for a dataset (Globus, SEAD, Dataverse) with a button/link to the "Job Submission" UI
- Job Submission UI is basically the Tool manager or Jupyter tmpnb
- At the top (faintly) is a registry that resolves a dataset URL to it's location with mountable path
- (There was some confusion whether this was the dataset URL or dataset DOI or other PID, but now it sounds like URL – see example below)
- On the right are the datasets at their locations (SDSC, NCSA)
- The user can launch a container (e.g., Jupyter) that mounts the datasets readonly and runs on a docker-enabled host at each site.
- Todo list on the right:
- Data access at SDSC (we need a docker-enabled host that can mount the Norman dataset)
- Auth – how are we auth'ing users?
- Container orchestration – how are we launching/managing containers at each site
- Analysis?
- BW → Condo : Copy the MHD dataset from Blue Waters to storage condo at NCSA
- Dataset metadata (Kenton)
- Resolution (registry) (Kyle)
Planning Discussion 2
Notes from discussion (Craig W, David R, Mike L) based on above whiteboard diagram:
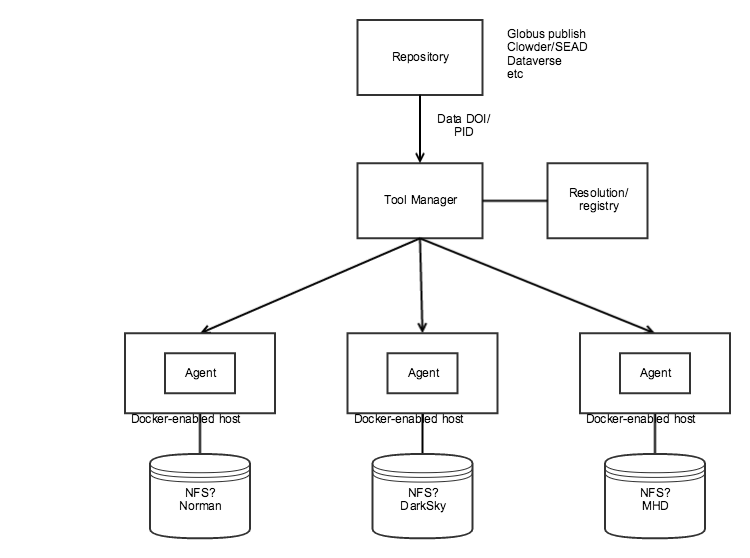
Components that we already have available:
- NCSA "Tool Manager" demonstrated at NDSC6
- Angular UI over a very simple Python/Flask REST API.
- The REST API allows you to get a list of supported tools, post/put/delete instances of running tools.
- Fronted with a basic NGINX proxy that routes traffic to the running container based on ID (e.g., http://tooserver/containerId/)
- Data is retrieved via HTTP get using repository-specific APIs. Only Clowder and Dataverse are supported
- Docker containers are managed via system calls (docker executable)
- WholeTale/ytHub/tmpnb:
- Girder with yt extension to support launching tmpnb notebooks
- tmpnb proxy and custom notebook server (volman) to support volume mounts
- PRAGMA PID service
- Demonstrated at NDSC6, appears to allow attaching arbitrary metadata to registered PID.
- Analysis
- For the Dark Sky dataset, we can use the notebook demonstrated by the yt team.
- For the Dark Sky dataset, we can use the notebook demonstrated by the yt team.
What we need to do:
- Copy MHD dataset to storage condo
- Create Docker-enabled hosts with access to each dataset (e.g., NFS) at SDSC, possibly in the yt DXL project, and in the NDS Labs project for MHD
- Decide whether to use/extend existing Tool Manager, Girder/yt/tmpnb (or something else)
- Define strategy for managing containers at each site
- Simple: "ssh docker run -v" or use the Docker API
- Harder: Use Kubernetes or Docker Swarm for container orchestration. For example, launch a jupyter container on a node with label "sdsc"
- Implement the resolution/registry
- Ability to register a data URL with some associated metadata.
- Metadata would include site (SDSC, NCSA) and volume mount information for the dataset.
- The PRAGMA PID service looks possible at first glance, but may be too complex for what we're trying to do. It requires handle.net integration.
- Implement bookmarklet or browser extension: There was discussion of providing some bookmarklet javascript to link a data DOI/PID to the "tool manager" service
- Authentication:
- TBD – how do we control who gets access, or is it open to the public?
- In the case of Clowder/Dataverse, all API requests include an API key
- Analysis:
- Need to get notebooks/code to demonstrate how to work with the MHD and Norman data.
Example case for resolution (not a real dataset for SC16)
- A dataset has a Globus Publish landing page https://publish.globus.org/jspui/handle/ITEM/113
- This dataset has the URL
- This would map to Nebula:
- /scratch/mdf/publication_113
Planning Discussion 3
Notes from third set of discussions (Craig, David, Mike – with Slack guidance from Kacper).
Component Options
Looking at the above diagram, we see four categories of services:
- Repository: remote repository (e.g., Globus Publish, Dataverse, etc)
- Resolver: Given a dataset identifier, return metadata about (e.g., location) and maybe launch the notebook. "Data DNS"
- Agent: Services installed at each site to enable launching notebooks (e.g., proxy, notebook launcher, docker, etc)
- Data: Actual mounted data at each site
All combinations are possible, although some combinations will likely be easier to accomplish than others.
Open question: We've discussed the idea of federated services versus a centralized "resolver" service. See the "Federation Options" section for details.
"Repository" - User Frontend
Options:
user installs bookmarkletthis may be restricted in modern browsers... more research is necessary- Pros
- browser-agnostic
- Cons
- probably lots of learning involved here
- user must seek out and install this
- no notion of authentication (see below)
- more complex scenarios may not work in modern browsers
- Pros
- user installs browser extension
- Pros
- more secure than bookmarklets... I guess?
- Cons
- probably lots of learning involved here
- user must seek out and install this
- no notion of authentication (see below)
- browser-specific (we would need to develop and maintain one for each browser)
- Pros
- developer(s) add a link to repo UI which leads to the existing ToolManager UI landing page, as in the NDSC6 demo
- Pros
- user does not need to install anything special on their local machine to launch tools
- most repos inherently have a notion of "user" whose username and/or email we can use to identify tools launched by this user
- Cons
- repo UI developers who want to integrate with us need to add one line to their source to integrate with us
- Dataverse, Clowder, Globus Publish, etc
- repo UI developers who want to integrate with us need to add one line to their source to integrate with us
- Pros
"Resolver" - API endpoint to resolve identifiers (e.g., DOI, URN, URL) to notebook URLs
Options:
- Extend the existing NCSA ToolManager to add a /lookup endpoint
- Pros
- Easy to set up and modify as we need to
- Cons
- Likely not a long-term solution, but simple enough to accomplish in the short-term
- Pros
- Girder+yt: add identifier to metadata and use mongo_search function to resolve
- Pros
- Well-documented, extensible API, with existing notions of file, resource, and user management
- Cons
- likely overkill for this system, as we don't need any of the file management capabilities for resolving
- language barriers in modifying Girder - python + javascript (raw? nodejs?)
- Pros
- Build REST API over etcd
- Pros
- Familiar - this is how the ndslabs API server works, so we can possibly leverage Craig's etcd.go
- Cons
- it might be quite a bit of work to build up a new API around etcd
- Pros
- PRAGMA PID service?
- Pros
- sounds remarkably similar to what we're trying to accomplish here
- supports a wide variety of different handle types (and repos?)
- Cons
- may be too complex to accomplish in the short term
- unfamiliar code base / languages
- Has specific notion of a PID that may be too restrictive.
- Won't support multiple location resolution?
- Pros
- Extend the existing NCSA ToolManager to add a /lookup endpoint
"Agent" - launches containers alongside the data on a Docker-enabled host
Options
- Use the existing ToolManager?
- Pros
- already parameterized to launch multiple tools (jupyter and rstudio)
- Cons
- no notion of "user" or authentication
- Pros
- Girder/tmpnb?
- Pros
- notebooks automatically time out after a given period
- inherited notion of "user"
- Cons
- can only launch single image type, currently (only jupyter)
- inherited notion of "user" may present an interesting auth problem - how do we share these accounts between sites?
- Girderisms: need to pass around "folderIds" or resolve dataset identifiers to folders.
- Pros
- Kubernetes / Docker Swarm?
- Pros
- familiar - this is how the ndslabs API server works, so we can possibly leverage Craig's kube.go
- orchestration keeps containers alive if possible when anything goes wrong
- Cons
- may be too complex to accomplish in the short term
- Pros
- docker -H?
- Pros
- zero setup necessary, just need Docker installed and the port open
- Cons
- HIGHLY insecure - would require some form of authentication (see below)
- Pros
- Use the existing ToolManager?
"Data" - large datasets need to be mountable on a Docker-enabled host
- NFS?
- GFS?
- S3?
- other options?
Federation options
- Centralized
- All datasets are registered with a central service that is responsible for resolving identifiers to locations and launching notebooks at those locations.
- Pros
- synchronization / authentication (see below) may be slightly easier to solve (only one user)
- Can mix data from two sites: having information about data in one place allows users to compose freely.
- Cons
- single point of failure
- Local access to datasets at a site requires going through an external service
- Federated
- Each site has it's own local stack but register with a federation server for id → location resolution
- Pros
- Users at each site can access data directly/launch notebooks without federation server
- Can use existing services, such as hub.yt
- Cons
- synchronization (see below) is still an open question (is this an open broadcast? handshake? do we keep a record of nearest neighbors?)
- authentication (see below) and sharing credentials between sites becomes a more complex problem
- Can't mix data from two sites
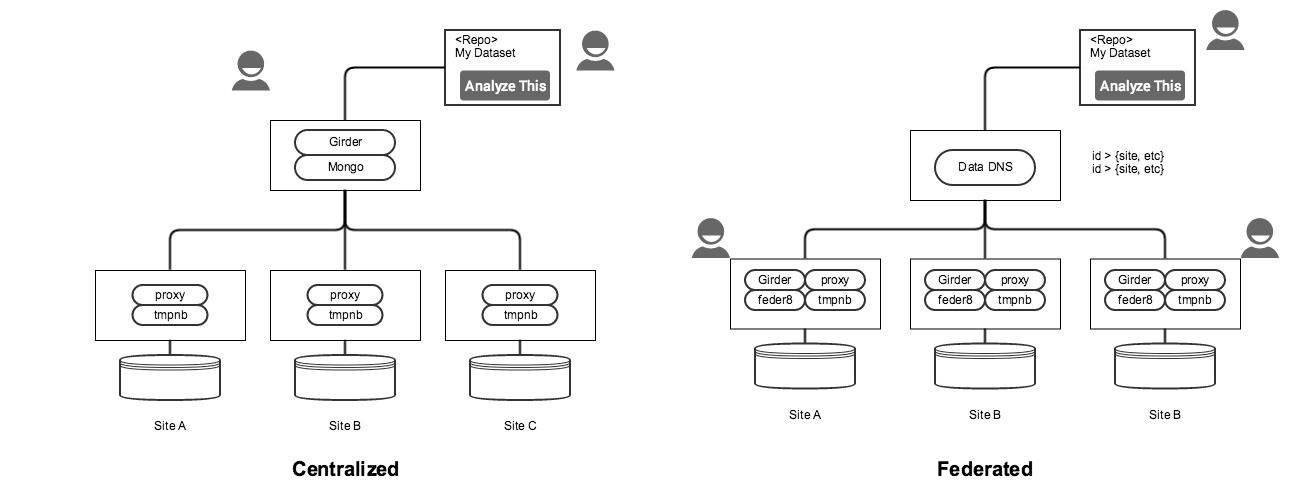
Additional notes for this diagram:
- Centralized
- Assuming that we use Girder as-is, the centralized model requires mounting each dataset filesystem via NFS/SSHFS for the initial metadata "ingest". This is only temporary and does not ingest the actual file data, but is awkward.
- We would need to use or extend the Girder API to support the remote repository request – resolving the dataset identifier (DOI, URL, URN) to the Girder folderId to get the notebook.
- Requires a user account on the Girder instance to launch notebooks at each site
- Solves the Whole Tale problem of running remote docker containers.
- Federated:
- New "Data DNS" component to handle registration and resolution of IDs to sites
- New "Federate" component at each site is needed to post data to the federation/Data DNS service
- Assumes local user accounts at each site – which means users can access the datasets without the federation server, but also means that there are unique user accounts at each site. Using tmpnb, we can't have a single guest user, since there's one notebook per user?
- In this model, we could use the hub.yt infrastructure as-is, with the addition of the "federat8" component. No need to copy or mount the DarkSky dataset.
- Doesn't solve the Whole Tale problem of running remote docker containers
Synchronization options
- Sites push their status to the resolver API
- Assumption: failures are retried after a reasonable period
- Pros
- Updates happen in real-time (no delay except network latency)
- Cons
- Congestion if many sites come online at precisely the same second
- More work for whatever we choose as the scheduler / orchestration system - a site missing a scheduled push means we may need to pull it out of rotation
- Resolver service polls for each site's status
- Assumption: failures are silent, and retried on the next poll interval
- Pros
- We will know explicitly when sites' are no longer available for launching tools
- Cons
- Time delay between polls means we could have stale data
- Threading nightmare - this is either one thread short-lived per site, or one giant thread looping through all sites
Authentication options
Auth may inhibit the "one-touch" behavior desired for these tools - you will always need to at least choose a tool and possibly enter credentials when launching a tool
- Build some kind of quasi-auth scheme (similar to ndslabs) on top of the existing ToolManager
- Inherit Girder's auth scheme and solve the problem of sharing these "users" between sites
- Create a "guest" user at each site and use that to launch tools from remote sources
- NOTE: tmpnb only allows one notebook per user (per folder?), so anyone launching remotely would be sharing a notebook
- this is undesirable, as ideally each request would launch a separate instance
- lingering question: how do we get you back to the notebook if you lose the link? how do we know which notebook is yours?
Inclinations: SC16 Demo
- Transfer (if necessary) each dataset to existing cloud architecture - in progress?
- Spin up a Docker-enabled host and mount nearby datasets (NFS, direct mount, etc.) - in progress?
- Federated model
- Using docker-compose, bring up provided girder-dev environment on each Docker host - pending
- Develop the "resolver" REST API to
- Receive site metadata in - done => POST to /datasets
- Delegate tmpnb requests to remote Girder instances using existing /notebook API endpoint - done => POST to /resolve/:id
- Add authentication:
- We simply need to collect an e-mail address (identity) to run things on their behalf
- Could possibly import existing users from Girder using their API? probably not, due to security
- We could callback to Girder when sites push their metadata (assuming this can be done as Girder comes online)
- Extend UI to list off collections in connected Girder instance - doneish... very primitive, but styling it is trivial
- Add a "Launch Notebook" button next to each dataset where no notebook is running - doneish... prototype is working, once real metadata is in place this is trivial
- Add a "Stop Notebook" button next to each dataset where a notebook has been launched - TBD
- this is a slightly tougher problem, as we now need to call out to every Girder's /notebook endpoint
- Modify girder-dev to POST site metadata on startup (feder8)- in progress
- Run a centralized resolver instance on Nebula for the purposes of the demo
Using the above we would be able to show:
- Searching for data on compute-enabled systems (albeit in a list of only 3 datasets registered in the system), possibly linking back to the original data source
- Launch a Jupyter notebook next to each remote dataset without explicitly navigating to where that data is stored (i.e. the Girder UI)
- How to bring this same stack up next to your data to make it searchable in our UI (we could even demonstrate this live, if it goes smoothly enough)
Inclinations: As a Long-Term service
- leverage existing Labs/Kubernetes API for authentication and container orchestration / access across remote sites
- etcd.go / kube.go can likely take care of talking to the necessary APIs for us, maybe needing some slight modification
- possibly extend Labs apiserver to include the functionality of delegating jobs to tmpnb and/or ToolManager agents?
- this leaves an open questions: single geodistributed kubernetes cluster? or one kubernetes cluster per site, federated across all sites ("ubernetes")
Notes from 10/28 meeting
Present: Mike, David, Kenton, Kandace, Kacper, Craig
- Kacper explained more of the Whole Tale design:
- There will be a website where the user can enter the DOI, DOI will resolve to remote repository (e.g., DataOne). Ingest will only happen at that point (on-demand)
- When the data can't be moved, if compute near the data it will be used
- Need to support composing multiple datasets – e.g., DarkSky + some smaller dataset. In this case, the smaller data will be moved to site with the large dataset.
- Might look into Agave project for capabilities API (long-term)
- Specific comments about SC16 demo:
- folderId requirement in volman can be removed – just hardcode the mountpoint. So can the userId requirement.
- tmpnb can be used to simply create a temporary notebook – not tied to a user/folder
- The folderId is useful when the user want's access to a subset/subdirectory
- tmpnb is nothing new, so alone this isn't much of a demo.
- DarkSky data is NFS mountable read
- Regarding Norman dataset, Girder does support Swift for ingest, but need to test it.
- Girder supports Oauth, if useful
- folderId requirement in volman can be removed – just hardcode the mountpoint. So can the userId requirement.
- There is now a presentation on November 16th
- Next steps – for the demo, we will used the "Federated" model above, but long term there's still much to discuss
- Data transfer (MHD)
- Write the registry API and UI for proof-of-concept
- Swift problem: Ingest into Girder directly or find out how best to mount Swift into container
- Create VM near data at SDSC with Girder stack
- Example notebooks for 3 datasets
- Discussion of big-data publishing stack (spitballing)
- Girder+tmpnb is now an option we can recommend to SCs to make these big datasets available. Install these services, and you can make an inaccessible dataset accessible, with minimal analysis support.
- This isn't the only stack – one of many options, but this works for the large physics simulation data.
- If they install this stack, they could (potentially, with much more thought) be Whole-Tale compatible.
- Discussion of "Data DNS" service (spitballing)
- This came up during an earlier whiteboard session. The resolver can be a sort of data DNS service – given an identifier, resolve to one or more locations.
- This would be different than the RDA PID concept, not an authoritative registry, just a way of saying "I have a copy of this data available here already" for larger datasets
- Sites could possibly publish capabilities – I have this data and can launch a Docker container (e.g., Jupyter); I have this data in Hadoop/HDFS; I can support MPI jobs, etc.
- The identifier now is anything that uniquely identifies the dataset (PID, DOI, Handle, URN, URL)
Storyboard for Demo Presentation
Here is my PPT version of my napkin sketch for the SC demo. Also context on where the demo product fits in the story. Comments, please!
Presentation v1
Please give me your feedback! Graphics executed to the edge of my abilities and patience. See presentation notes for script written so far.