Resources
BioCADDIE project overview:
- BioCADDIE challenge: modeled after Text REtrieval Conference (TREC)
- Benchmark dataset
- 15 test queries
- Metrics for evaluation
What did we do?
- Pseudo-relevance feedback (query expansion) using an outside collection (PubMed Open Access collection)
- Repository prior
What is pseudo-relevance feedback (PRF):
- Issue the query; treat the top N documents as relevant; expand the query using the top terms from the top documents; rescore/resubmit query
- It's usually effective, but there's en effectiveness/efficiency tradeoff.
What will we deliver:
- Package and document what we did for the challenge (Craig)
- Re-evaluate models with full test collection
- Include document expansion
- Include other external collections (e.g., Wikipedia)
- Choose best model
- Implement model in current pipeline
- PubMed ingest (if useful)
- Implement Rocchio or RM in Lucene (or find existing)
Specific tasks
| Task | Status | |
|---|---|---|
| Pre-development | Package/document existing code | Done |
| Infrastructure | Get SDSC Cloud Allocation | Done |
| Provision development VMs | Done | |
| Development environment | Get Access to DataMed source | N/A |
| Get DataMed pipeline up and running on SDSC | N/A | |
| Comparative evaluation | Re-run baselines on final test collection | Done |
| Re-run document expansion tests | Done | |
| Re-run RM1 and RM3 tests | Done | |
| Run Rocchio tests | Done | |
| Run sequential dependence model | Done | |
| Final comparison/report against challenge results | ||
| Development | Determine best way to add feedback to ElasticSearch | Done |
| Determine best way to add prior to ElasticSearch | ||
| PubMed ingest and maintenance process | In-progress | |
| Implement document expansion process | Won't do | |
| Implement query expansion API | Done | |
| Implement repository prior | ||
| Testing | Test plan including functional, integration, and performance tests | Not done |
| Execute test plans | Not done | |
| Release | Release planning, packaging | Not done |
| Documentation | Document components, architecture, installation, administration | In progress |
Brief overview of information retrieval concepts:
For this project, we are primarily concerned with what is called "ad-hoc" information retrieval (aka search). This simply means that users enter arbitrary queries against an index of documents, as we're all accustomed to for web search. Below are a few key concepts that you could be familiar with for this project.
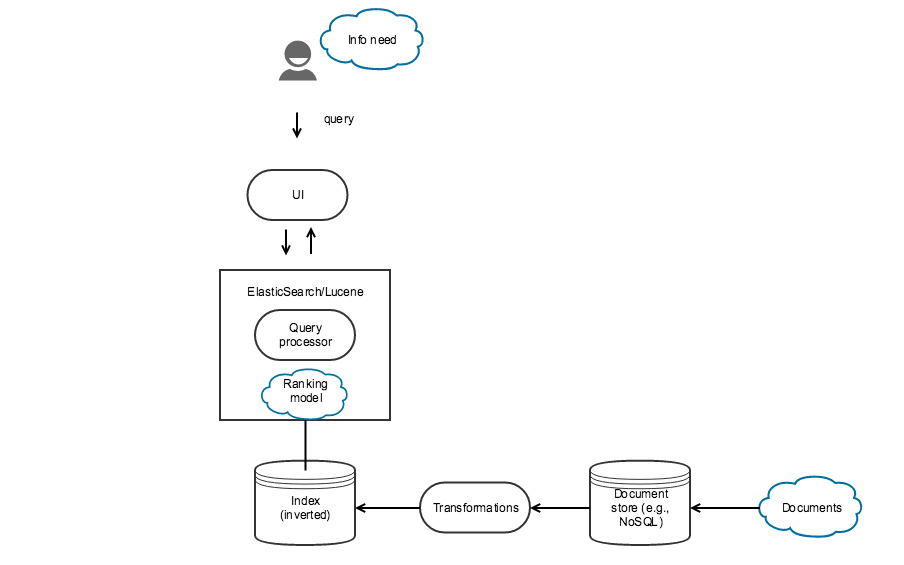
Search engine architecture
- Text acquisition: Some means of getting the documents to be indexed (e.g., crawling the web)
- Document storage: A system to store the raw documents. These days this is often a NoSQL system or something like BigTable
- Text transformation: Subsystem to transform text prior to indexing. Common processes include stopping, stemming, expansion, link analysis, information extraction, classification, named entity recognition.
- Index creation: Process to actually create/update the index based on the document collection.
Search engines:
Queries and information needs
- Information need: the underlying cause of the query. This is latent, unobservable.
- Query: the text the user enters into the search box. The same query can represent multiple information needs; the same information need may be represented by multiple queries. This is the observable thing that we can use to actually compare to documents.
Ranking and similarity
- Relevance: Abstract concept used to describe the judgement of whether a particular document fulfills or does not fulfill the user's information need.
- Similarity: How we operationalize relevance. Similarity may be operationalized geometrically (e.g., vector space models), probabilistically (e.g., language models), logically (e.g., Boolean).
- Ranking models: There are a number of different ranking models or scoring algorithms available in search engine software. Common models include Boolean, TFxIDF, BM25, and language models.
Two broad classes of models:
- Vector space: Treat documents/queries as vectors in n-dimensional space. Measure angle between as "similarity"
- Probabilistic (language modeling): Treat documents/queries as probability distributions over terms. Use probabilistic models to measure "similarity" – e.g., P(Q | D).
See also:
- ElasticSearch documentation on similarity modules
- Search Engines: Information Retrieval in Practice by W.B. Croft, D. Metzler, T. Strohman
- Statistical Language Models for Information Retrieval by ChengXiang Zhai
- A General Theory of Relevance by Viktor Lavrenko
Evaluation
How do we know whether one model is better than another?
- Test collection:
- A set of documents
- A set of queries
- Relevance judgments (query/document pairs, generally pooled)
- Metric(s)
Metrics:
- Precision, recall, mean average precision, precision@k, normalized discounted cumulative gain (NDCG)
Feedback and expansion models
- Queries are often too sparse and have vocabulary mismatch with documents
- Query expansion helps to alleviate the problem.
- Relevance feedback: User's judge document relevance, use judged-relevant documents to expand queries
- Psuedo-relevance feedback: Assume top K documents are relevant, use to expand query.
- Two commonly used models
- Rocchio (vector space framework)
- Lavrenko's relevance models (language modeling framework)
See also: