8/31/2017
From 2017-08-31 Workbench planning, we'll consider the following use cases:
- Running ETK on Comet from WB
- TERRA-REF, either stitching or Genomics workflows
- Other: TERRA Fusion, LSST
8/3/2017
Brainstorming:
- Use case:
- HPC operator installs WB to provide:
- Interactive analysis/development environments with access to shared data
- Access to Hadoop and traditional TORQUE/PBS resources (headnode replacement)
- WB is just 1+ nodes in the cluster with access to shared filesystem, authentication, etc.
- User
- Develops in WB, creates and deploys containers (Docker, Singularity) to local registry (or Dockerhub)
- Writes PBS/qsub scripts and submits job from within WB, can monitor progress. Similar for Hadoop.
- Data is available to WB for analysis.
- HPC operator installs WB to provide:
- Benefits:
- User has one-stop-shop for development, exploratory analysis, visualization and data access that isn't the head node
- HPC operator no longer needs to provide access to dedicated head node. Users can launch custom software without requiring additional installation.
- HPC operator can easily support running workshops/tutorials etc.
- Essentially, users would have qsub/qdel/qstat all available in containers in WB instead of SSHing into the head node.
- We could do this easily via SCP/SSH (ssh clusterserver qsub /tmp/foo.sh), very easy if we share the home directory.
- Could also develop (or leverage existing?) method for remote job submission API (Agave?)
8/2/2017 Notes (Craig, Mike)
Discussed the SC17 demo as something that's also demo'd at NDS8 and is part of the vision for the SI2 proposal.
TERRA-REF use case:
- ROGER Supercomputer hosts the TERRA-REF project data
- ROGER has OpenStack, HPC (TORQUE) and Hadoop support
- Data is stored on GPFS, which can be mounted via NFS
- TERRA-REF Workbench already mounts the TERRA ROGER data
Demo workflow 1: From zero to HPC
- Develop some idea in WB using the TERRA-REF data
- Run/test in WB on subset of TERRA data
- Build code into Docker image in WB
- Run analysis on TORQUE or Hadoop clusters
Demo workflow 2: From zero to Cloud
- Start some service in WB (e.g., Clowder)
- Do your customization
- Export to Docker compose (link to zip file containing AppData and docker compose file)
- On OpenStack, AWS or GCE VM with Docker support, wget and unzip, then docker-compose up. Viola – your service.
Running jobs on ROGER:
- https://wiki.ncsa.illinois.edu/display/ROGER/ROGER+User+Guides
- ssh you@roger-login.ncsa.illinois.edu
qsub -l nodes=1:ppn=1,walltime=600 -o myjob.log myjob.pbs
- qstat <jobid>
- qdel <jobid>
Questions:
- Yan suggested adding support for NCSA LDAP/PAM authentication to Workbench. This way, UID/GID match ROGER.
- Could possibly mount ROGER home directory on Workbench?
- Starting jobs on ROGER seems to require qsub, period. Will talk with Yan, but there's no apparent API.
- What about an SSH tunnel to ROGER?
- Or adding something to authorized hosts, if the user directory can be mounted?
- pbs_server seems to have a host/port. Perhaps we can configure qsub in WB to talk to pbs_server?
July notes
We've discussed the TERRA-REF hyperspectral workflow as a possible example:
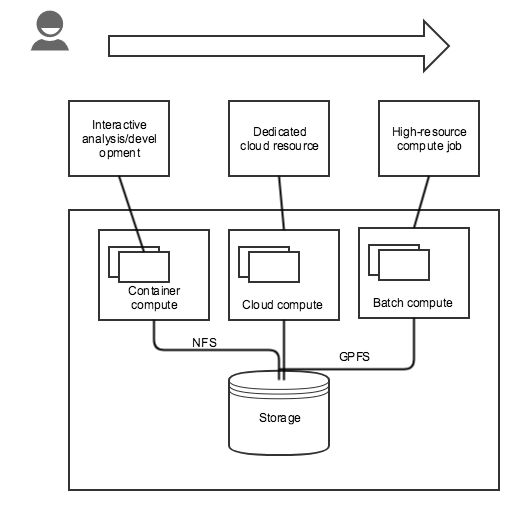
In the case of the hyperspectral extractor development, the hyperspectral data is too large to move so the researcher/developer working on related algorithms needs to work on data in place. This is done via a NetCDF/NCO-specific development environment, but could just as easily by Jupyter or RStudio. If the researchers needs exceed container constraints, they can apply for dedicated cloud resources via OpenStack. They also have access to run jobs on ROGER via PBS. This is similar to Cyverse.
Fruend Case (CSE UCSD, Kevin)
- Data analysis using Juypter Notebooks and Spark for the student Capstone Projects in the MAS Data Science and Engineering program.
- Launches Spark Clusters using Elastic Map Reduce (EMR) on AWS.
- When the EMR cluster is created a bootstrap script is passed to the cluster to install and configure Juypter.
- Uses the Flask Python framework to launch a locally running web server to make it easy to configure AWS credentials and launch the EMR Cluster.
- Students do work in the Jupyter notebooks by connecting to the EMR SparkContext using pyspark.
- Python libraries are included that make it easy to copy data to/from S3, HDFS and the local filesystem on the Spark master.
- https://github.com/mas-dse/spark-notebook